Service
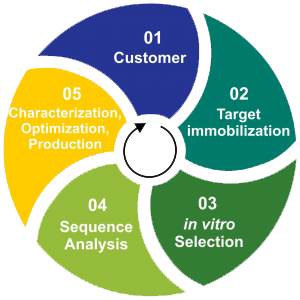
Our selection service includes an automated selection of RNA Aptamers against protein targets. The standard automated selection contains, target immobilization, aptamer selection, and characterization with the corresponding SOPs. Sequence analysis and aptamer optimization are optional and will be discussed with CARD users. Besides the standard automated selection, we also offer an optional support as a post-selection phase, which includes Next Generation Sequencing (NGS), as well as affinity and specificity determination of the resultant aptamers.We are happy to provide you with additional information about our services.
Please use our formular to contact us.
CARD’s proprietary platform facilitates the automated selection of aptamers, able to perform twelve iterative selection cycles without manual interference. The current aptamer selection protocol at CARD is designed for the generation of RNA and 2’Deoxy-2’Fluoro Pyrimidine RNA aptamers for protein targets. However, protocols for covering other targets, e.g. cells or small molecules and nucleic acid types, e.g. DNA and nucleobase-modified DNA will be implemented and available soon. The automated selection process enables the identification of enriched libraries within a short timeframe. A summary of recently developed aptamers using CARD’s fully automated platform is listed in table 1.
| Target | Nucleic Acid | Reference |
|---|---|---|
| Erk1/2 | RNA | Lennarz, S. et al. Angew. Chem 2015 Lennarz, S. et al. ACS Chem Biol 2015 |
| GRK2 | RNA | Mayer et al. RNA 2008 Tesmer et al. Structure 2012 |
| Lysozyme | RNA | Unpublished |
Table 1: RNA aptamers developed using CARD’s fully automated selection platform.